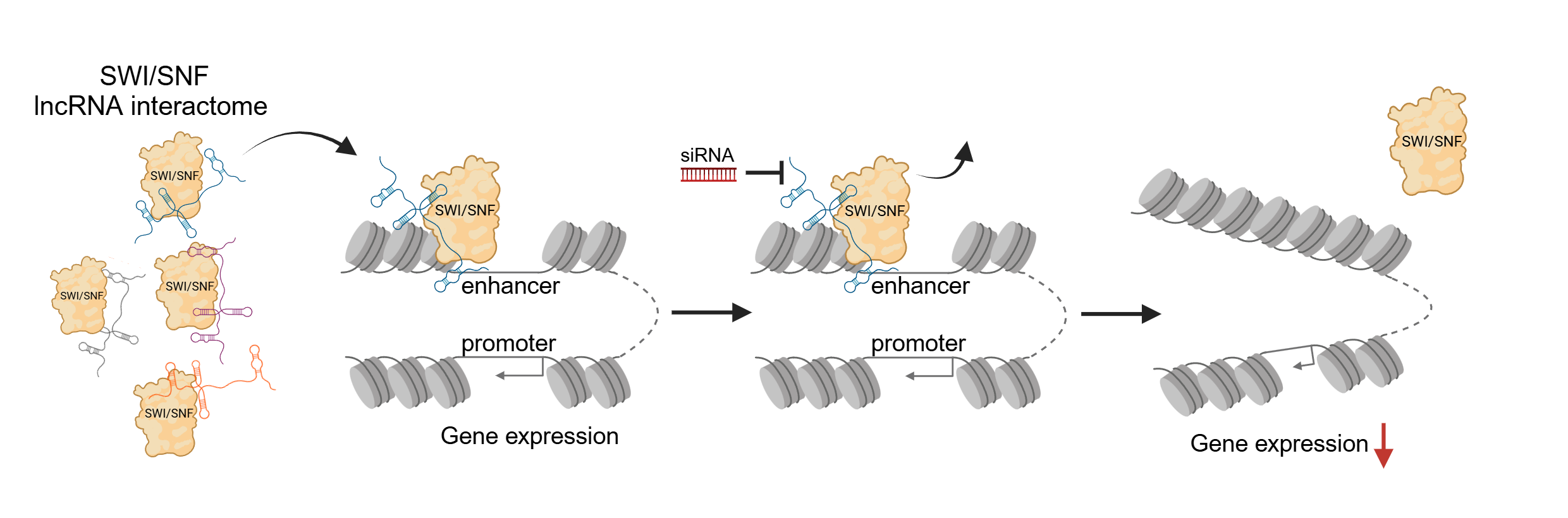
Chromatin remodeling is essential for DNA accessibility and gene expression control in health and disease. How such a fundamental process is coordinated in a spatial and temporal manner remains unknown. The SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complex is expressed in every cell type from yeast to humans and remodels chromatin genome-wide. In particular, SWI/SNF binds lineage specific enhancers where it actively maintains open chromatin state while being able to respond dynamically to cellular signals.
The mechanisms that guide SWI/SNF to specific genomic targets have remained elusive. Here Dr. James Alexander Oo and his colleagues from Matthias Leisegang`s group have demonstrated that trans-acting long non-coding RNAs (lncRNAs) direct the SWI/SNF complex to cell type-specific enhancers. They performed a technique called RedChIP which allowed for the pulldown of SWI/SNF proteins bound to both RNA and DNA. Sequencing of the SWI/SNF-enriched RNA-DNA pinpointed exactly which lncRNA was bound to which DNA site with SWI/SNF.
After the validation of interactions with techniques such as iCLIP and CUT&RUN, they perfomed a knockdown screen against the candidate lncRNAs. This led to the genome-wide redistribution of SWI/SNF away from specific enhancers and a concomitant differential expression of spatially connected target genes.
These findings reveal that lncRNAs competitively recruit SWI/SNF, providing a specific and dynamic layer of control over chromatin accessibility, and reinforcing their role in mediating enhancer activity and gene expression. Considering that SWI/SNF is fundamental in differentiation and is mutated in 25% of cancers, a better understanding of how it is recruited in a cell type- and context-dependent manner offers huge therapeutic potential.
Find the full article here:
You need to load content from hCaptcha to submit the form. Please note that doing so will share data with third-party providers.
More InformationYou need to load content from reCAPTCHA to submit the form. Please note that doing so will share data with third-party providers.
More InformationYou need to load content from reCAPTCHA to submit the form. Please note that doing so will share data with third-party providers.
More InformationYou need to load content from reCAPTCHA to submit the form. Please note that doing so will share data with third-party providers.
More InformationYou need to load content from Turnstile to submit the form. Please note that doing so will share data with third-party providers.
More Information